Include dependency graph for kmeans_landmark.h:
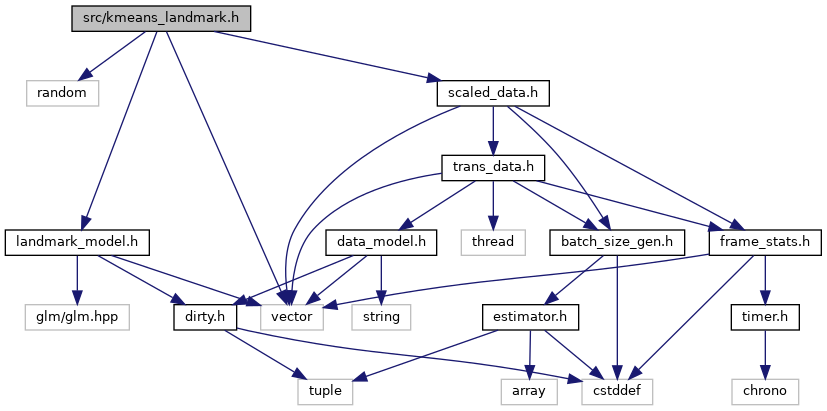
This graph shows which files directly or indirectly include this file:
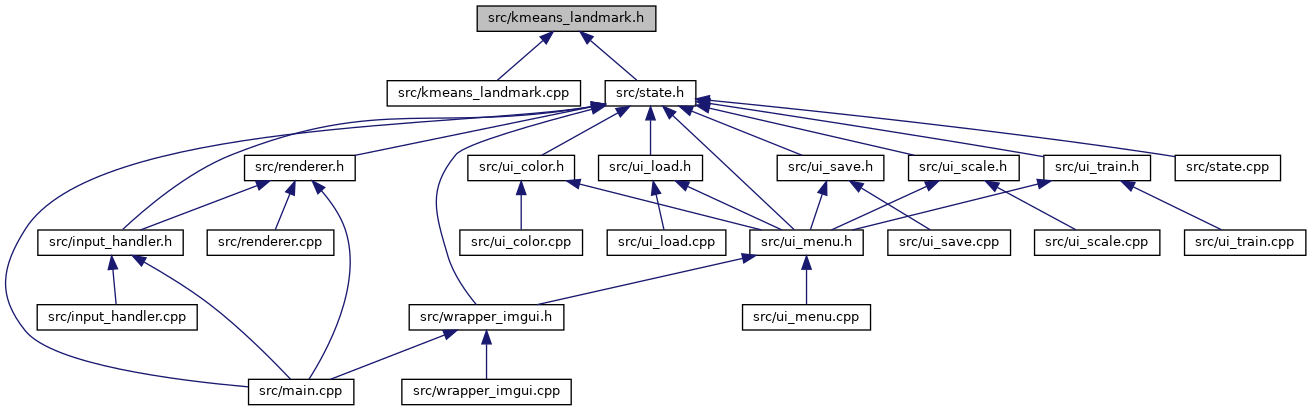
Go to the source code of this file.
Classes | |
| struct | KMeansData |
| Structure for storing the kmeans-style data. More... | |
Functions | |
| void | kmeans_landmark_step (KMeansData &data, const ScaledData &model, size_t iters, float alpha, float neighbor_alpha, LandmarkModel &lm) |
| Run a k-means-like optimization of high-dimensional landmark positions. More... | |
| void | som_landmark_step (KMeansData &data, const ScaledData &model, size_t iters, float alpha, float sigma, LandmarkModel &lm) |
| Run a SOM to optimize high-dimensional landmark positions. More... | |
Function Documentation
◆ kmeans_landmark_step()
| void kmeans_landmark_step | ( | KMeansData & | data, |
| const ScaledData & | model, | ||
| size_t | iters, | ||
| float | alpha, | ||
| float | neighbor_alpha, | ||
| LandmarkModel & | lm | ||
| ) |
Run a k-means-like optimization of high-dimensional landmark positions.
Definition at line 36 of file kmeans_landmark.cpp.
Here is the call graph for this function:
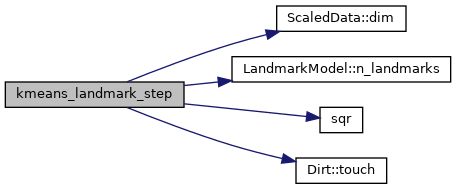
Here is the caller graph for this function:

◆ som_landmark_step()
| void som_landmark_step | ( | KMeansData & | data, |
| const ScaledData & | model, | ||
| size_t | iters, | ||
| float | alpha, | ||
| float | sigma, | ||
| LandmarkModel & | lm | ||
| ) |
Run a SOM to optimize high-dimensional landmark positions.
Definition at line 87 of file kmeans_landmark.cpp.
Here is the call graph for this function:
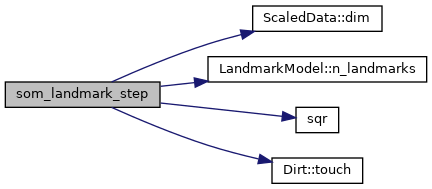
Here is the caller graph for this function:
